R is a powerful language used widely for data analysis and statistical computing. Inclusion of powerful packages in R has made it more and more powerful with time. One such recent and wonderful package for Covid-19 is ‘covid19.analytics‘.
COVID19.Analytics Package
Introduction
The “covid19.analytics” R package allows users to obtain live worldwide data from the novel Corona Virus Disease originally reported in 2019, Covid-19, as published by the JHU CCSE repository, as well as, provide basic analysis tools and functions to investigate these datasets.
The goal of this package is to make the latest data promptly available to researchers and the scientific community. The package is really handy when getting and visualizing real time COVID data. It provides daily count of cases,recoveries and deaths due to this virus. The predefined functions are even more useful. It has inbuilt GLM models for covid and even genome sequence for viruses. It also includes SIR model which actually shows the importance of social distancing to slowing the spread of the virus.
How to Install this package?
install.packages(“covid19.analytics”)
Loading Package in R
library(covid19.analytics)
Overview of the main functions from the “covid19.analytics” Package:
- covid19.data: This function is used to read “live” data from reported Covid’19 cases.
mydata = covid19.data(case = “aggregated”, local.data = FALSE, debrief = FALSE)
Arguments:
case:
| Argument | description |
Aggregated | latest number of cases aggregated by country |
| Time Series data | |
ts-confirmed | time series data of confirmed cases |
ts-deaths | time series data of fatal cases |
ts-recovered | time series data of recovered cases |
ts-ALL | all time series data combined |
| Deprecated data formats | |
ts-dep-confirmed | time series data of confirmed cases as originally reported (deprecated) |
ts-dep-deaths | time series data of deaths as originally reported (deprecated) |
ts-dep-recovered | time series data of recovered cases as originally reported (deprecated) |
| Combined | |
ALL | all of the above |
| Time Series data for specific locations | |
ts-Toronto | time series data of confirmed cases for the city of Toronto, ON - Canada |
ts-confirmed-US | time series data of confirmed cases for the US detailed per state |
ts-deaths-US | time series data of fatal cases for the US detailed per state |
local.data : boolean flag to indicate whether the data will be read from the local repo, in case of connectivity issues or data integrity
debrief : boolean specifying whether information about the read data is going to be displayed in screen
Data Structure:
The Time Series data is organized in a specific manner with a given set of fields or columns, which resembles the following structure:
View(mydata)
Using your own data and/or importing new data sets:
If you have data structured in a data.frame organized as described above, then most of the functions provided by the “covid19.analytics” package for analyzing Time Series data will work with your data. In this way it is possible to add new data sets to the ones that can be loaded using the repositories predefined in this package and extend the analysis capabilities to these new datasets.
names(mydata)
Be sure also to check the compatibility of these datasets using the Data Integrity and Consistency Checks functions.
Analytical & Graphical Indicators:
In addition to the access and retrieval of the data, the package includes some basics functions to estimate totals per regions/country/cities, growth rates and daily changes in the reported number of cases.
tsc = covid19.data(“ts-confirmed”)
- report.summary: This function is used to summarize the current situation, it will first download the latest data and then summarize the top provinces/cities per case. It results on screen table and static plots (pie and bar plots) with reported information. It can also output the tables into a text file.
report.summary(cases.to.process=”TS”, saveReport = FALSE, graphical.output = TRUE, geo.loc = NULL)
Arguments:
cases.to.process | which data to process: “TS” –time series–, “AGG” –aggregated– or “ALL” –time series and aggregated– |
Nentries | number of top cases to display (by default = 10 cases) |
geo.loc | geographical location to process |
graphical.output | flag to deactivate graphical output |
saveReport | flag to indicate whether the report should be saved in a file |
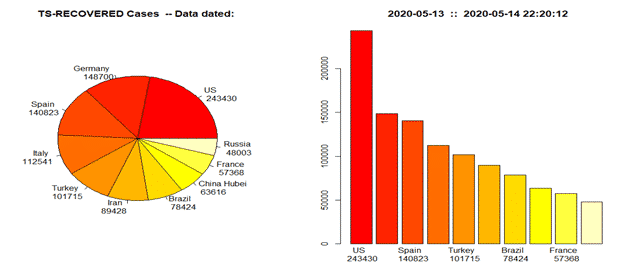
- tots.per.location : It compute totals per region and plot time series for that specific region/country. It provides static plots: data + models (exp/linear, Poisson, Gamma), mosaic and histograms when more than one location are selected.
tots.per.location(tsc, geo.loc =”India”, confBnd = FALSE, nbr.plts = 1, info=” “)
Arguments:
data | data.frame with *time series* data from covid19 |
geo.loc | list of locations |
confBnd | flag to activate/deactivate drawing of confidence bands base on a moving average window |
nbr.plts | parameter to control the number of plots to display per figure |
info | additional info to display in plots’ titles |
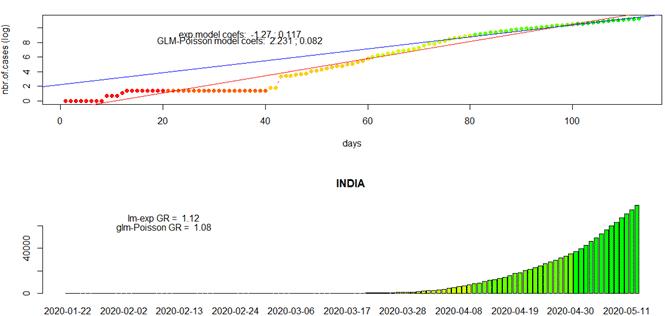
- growth.rate : It compute changes and growth rates per region and plot time series for that specific region/country. It displays list containing two dataframes: one reporting changes on daily basis and a second one reporting growth rates, for the indicated regions. It also produces static plots: data + models (linear,Poisson,Exp), mosaic and histograms when more than one location are selected.
growth.rate(data0=tsc, geo.loc =”India”, stride=1, info=””)
Arguments:
data | data.frame with *time series* data from covid19 |
geo.loc | list of locations |
confBnd | flag to activate/deactivate drawing of confidence bands base on a moving average window |
nbr.plts | parameter to control the number of plots to display per figure |
info | additional info to display in plots’ titles |
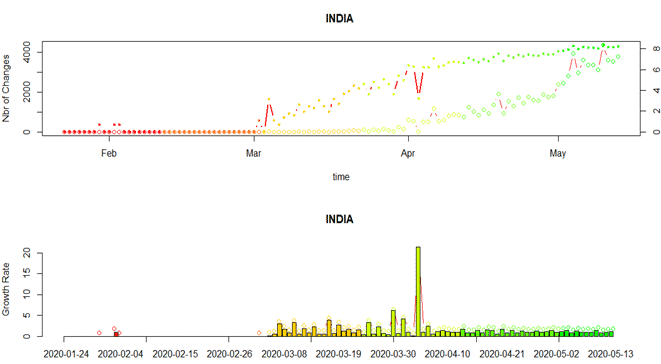
B) Growth Rate in Covid-19 cases in India on daily basis
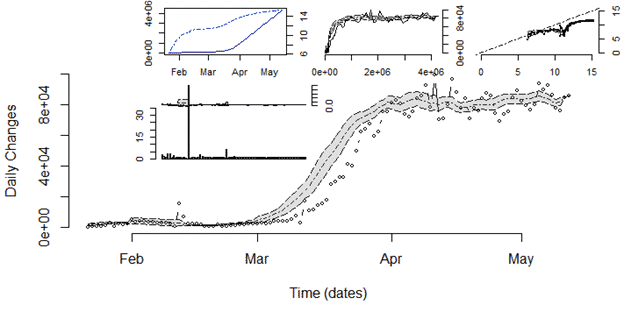
- single.trend: It is a function to visualize different indicators for trends in daily changes of cases reported as time series data. It is composed of static plots: total number of cases vs time, daily changes vs total changes in different representations.
single.trend(tsc, confBnd = TRUE, info = “”)
Arguments:
ts.data | time series data |
confBnd | optional argument to remove the drawing of a confidence band |
info | addtional information to display in plots |
Graphics and Visualization:
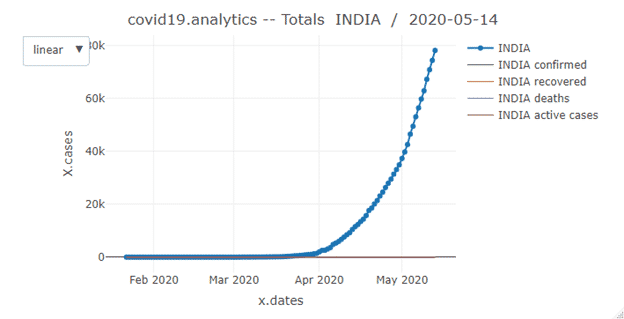
- total.plts: This function is used to plot total number of cases per day for different groups. It produces static and interactive plot
totals.plt(data0 = tsc, geo.loc0 =”India”, one.plt.per.page = FALSE, log.plt = FALSE, with.totals = FALSE, interactive.fig = TRUE, fileName = NULL)
Arguments:
data0 | time series dataset to process, default all the possible cases: ‘confirmed’ and ‘deaths’ for all countries/regions |
geo.loc0 | geographical location, country/region or province/state to restrict the analysis to |
one.plt.per.page | boolean flag to have one plot per figure |
log.plt | include a log scale plot in the static plot |
with.totals | a boolean flag to indicate whether the totals should be displayed with the records for the specific location |
interactive.fig | swith to turn off/on an interactive plot |
filename | file where to save the HTML version of the interactive figure |
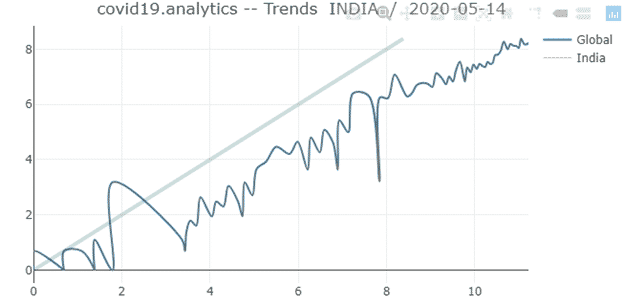
- itrends: It is a function to visualize trends in daily changes in time series data interactively.
itrends(ts.data =tsc, geo.loc =”INDIA”, with.totals = FALSE, fileName = NULL)
Arguments:
ts.data | time series dataset to process |
geo.loc | geographical location, country/region or province/state to restrict the analysis to |
with.totals | a boolean flag to indicate whether the global totals should be displayed with the records for the specific location |
fileName | file where to save the HTML version of the interactive figure |
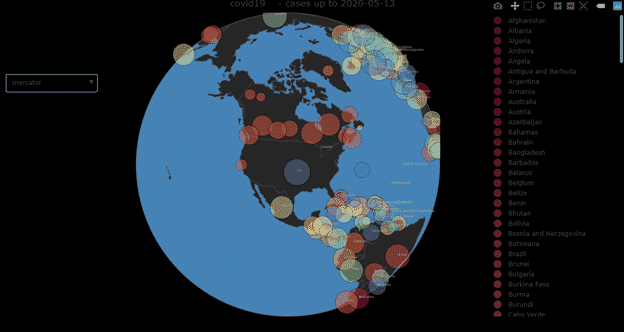
- live.map: It generates an interactive map displaying cases around the world.
live.map(data = tsc, select.projctn = TRUE, projctn = “orthographic”, title = “”, no.legend = FALSE, szRef = 0.2, fileName = NULL)
Arguments:
Data | data to be used |
| select.projctn | argument to activate or deactivate the pulldown menu for selecting the type of projection |
Projctn | initial type of map-projection to use, possible values are: “equirectangular” | “mercator” | “orthographic” | “natural earth” | “kavrayskiy7” | “miller” | “robinson” | “eckert4” | “azimuthal equal area” | “azimuthal equidistant” | “conic equal area” | “conic conformal” | “conic equidistant” | “gnomonic” | “stereographic” | “mollweide” | “hammer” | “transverse mercator” | “albers usa” | “winkel tripel” | “aitoff” | “sinusoidal” |
| Title | a string with a title to add to the plot |
| no.legend | parameter to turn off or on the legend on the right with the list of countries |
| szRef | numerical value to use as reference, to scale up the size of the bubbles in the map, from 0 to 1 (smmaller value –> larger bubbles) |
| Filename | file where to save the HTML version of the interactive figure |
Modelling:
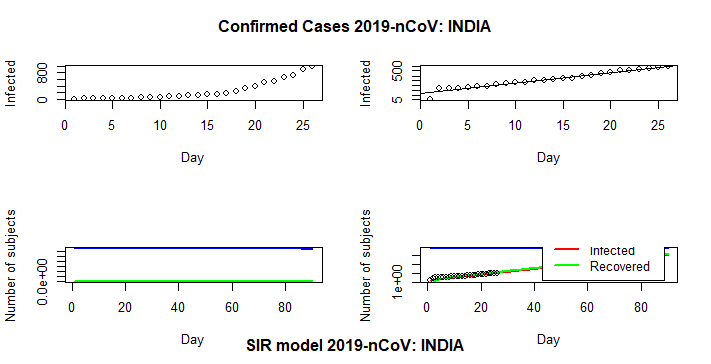
SIR (Suspectible – Infected – Recovered) Model:
An SIR model is an epidemiological model that computes the theoretical number of people infected with a contagious illness in a closed population over time. The name of this class of models derives from the fact that they involve coupled equations relating the number of susceptible people S(t), number of people infected I(t), and number of people who have recovered R(t).
| s(t) = S(t)/N, | the susceptible fraction of the population, |
| i(t) = I(t)/N, | the infected fraction of the population, and |
| r(t) = R(t)/N, | the recovered fraction of the population. |
where N is the total population
and at each time t, s(t) + i(t) + r(t) = 1
- generate.SIR.model: It generates a SIR (Susceptible-Infected-Recovered) model based on the actual data of the Covid-19 cases. It provides list containing the fits for the SIR model.
model = generate.SIR.model(data = tsc, geo.loc = “India”,
t0 = NULL, t1 = NULL, deltaT = NULL,
tfinal = 90, fatality.rate = 0.02, tot.population = 130*10^9,
staticPlt = TRUE, interactiveFig = FALSE)
Arguments:
data | time series dataset to consider |
geo.loc | country/region to analyze |
t0 | initial period of time for data consideration |
t1 | final period of time for data consideration |
deltaT | interval period of time from t0, ie. number of days to consider since t0 |
tfinal | total number of days |
fatality.rate | rate of causality, deafault value of 2 percent |
tot.population | total population of the country/region |
staticPlt | optional flag to activate/deactive plotting of the data and the SIR model generated |
interactiveFig | optional flag to activate/deactive the generation of an interactive plot of the data and the SIR model generated |
- plt.SIR.model: It is a function to plot the results from the SIR model function. It results static and interactive plots.
plt.SIR.model(SIR.model=model, geo.loc = “India”,
interactiveFig = FALSE, fileName = NULL)
Arguments:
SIR.model | model resulting from the generate.SIR.model() fn |
geo.loc | optional string to specify geographical location |
interactiveFig | optional flag to activate interactive plot |
fileName | file where to save the HTML version of the interactive figure |
Make sure you try these functions by yourself. You can just play with the arguments and come up with interesting analysis from the global live data available and thus develop a model which makes good prediction along with awesome graphics.
Have you used covid19.analytics package for your project? or Do you know of any other interesting package? I would love to hear from you! Connect with me in the comments section below or message me on LinkedIn and let’s talk R!

